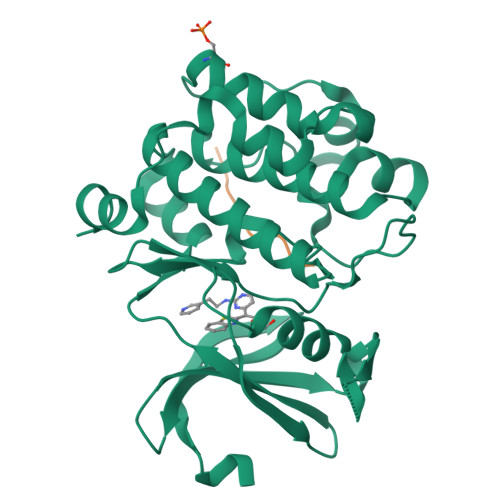
Proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V.
Filippakopoulos, P., Bullock, A., Fedorov, O., Pike, A.C.W., von Delft, F., Arrowsmith, C.H., Edwards, A.M., Bountra, C., Knapp, S.To be published.